Examples¶
Here we discuss some example systems and how to code them up using cayenne.
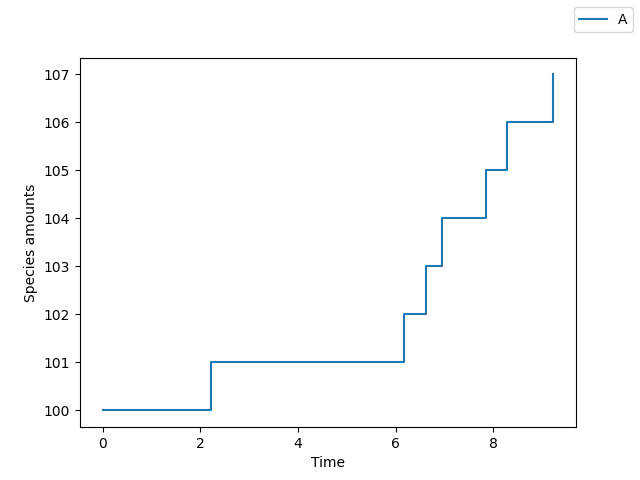
Zero order system¶
\[\begin{split}\phi &\xrightarrow[]{k_1} A\\
\\
k_1 &= 1.1\\
A(t=0) &= 100\\\end{split}\]
This can be coded up with:
>>> from cayenne import Simulation
>>> model_str = """
const compartment comp1;
comp1 = 1.0; # volume of compartment
r1: => A; k1;
k1 = 1.1;
chem_flag = false;
A = 100;
"""
>>> sim = Simulation.load_model(model_str, "ModelString")
>>> sim.simulate()
>>> sim.plot()
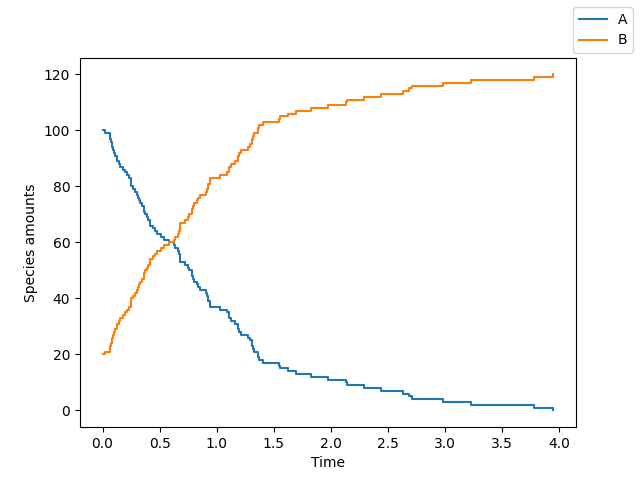
First order system¶
\[\begin{split}A &\xrightarrow[]{k_1} B\\
\\
k_1 &= 1.1\\
A(t=0) &= 100\\
B(t=0) &= 20\\\end{split}\]
This can be coded up with:
>>> from cayenne import Simulation
>>> model_str = """
const compartment comp1;
comp1 = 1.0; # volume of compartment
r1: A => B; k1;
k1 = 1.1;
chem_flag = false;
A = 100;
B = 20;
"""
>>> sim = Simulation.load_model(model_str, "ModelString")
>>> sim.simulate()
>>> sim.plot()
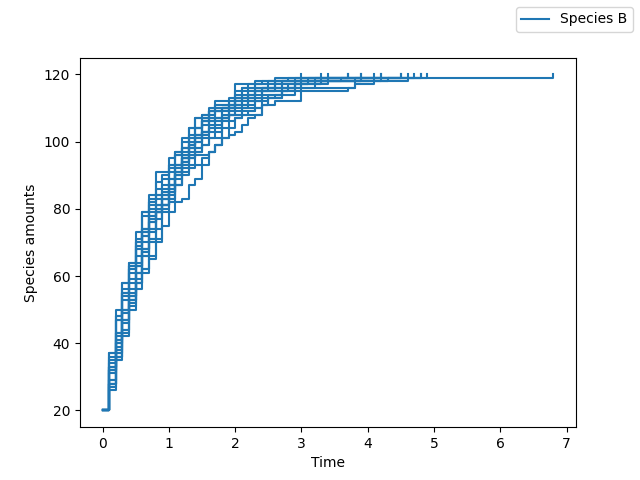
Suppose you want to use the tau_leaping algorithm, run 20 repetitions and plot only species \(B\). Then do:
>>> sim.simulate(algorithm="tau_leaping", n_rep=20)
>>> sim.plot(species_names=["B"], new_names=["Species B"])
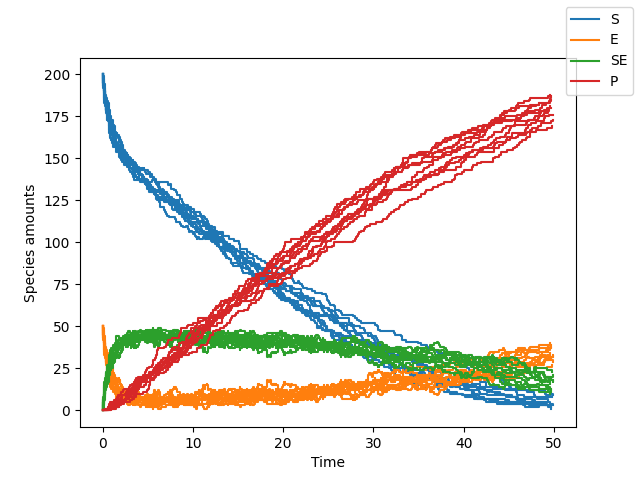
Enzyme kinetics (second order system with multiple reactions)¶
\[\begin{split}\text{Binding}: S + E &\xrightarrow{k1} SE \\
\text{Dissociation}:SE &\xrightarrow{k2} S + E \\
\text{Conversion}: SE &\xrightarrow{k3} P + E \\
\\
k1 &= 0.006 \\
k2 &= 0.005 \\
k3 &= 0.1 \\
S(t=0) &= 200\\
E(t=0) &= 50\\
SE(t=0) &= 0\\
P(t=0) &= 0\\\end{split}\]
This can be coded up with:
>>> from cayenne import Simulation
>>> model_str = """
const compartment comp1;
comp1 = 1.0; # volume of compartment
binding: S + E => SE; k1;
dissociation: SE => S + E; k2;
conversion: SE => P + E; k3;
k1 = 0.006;
k2 = 0.005;
k3 = 0.1;
chem_flag = false;
S = 200;
E = 50;
SE = 0;
P = 0;
"""
>>> sim = Simulation.load_model(model_str, "ModelString")
>>> sim.simulate(max_t=50, n_rep=10)
>>> sim.plot()
Since this is a second order system, the size of the system affects the reaction rates. What happens in a larger system?
>>> from cayenne import Simulation
>>> model_str = """
const compartment comp1;
comp1 = 5.0; # volume of compartment
binding: S + E => SE; k1;
dissociation: SE => S + E; k2;
conversion: SE => P + E; k3;
k1 = 0.006;
k2 = 0.005;
k3 = 0.1;
chem_flag = false;
S = 200;
E = 50;
SE = 0;
P = 0;
"""
>>> sim = Simulation.load_model(model_str, "ModelString")
>>> sim.simulate(max_t=50, n_rep=10)
>>> sim.plot()
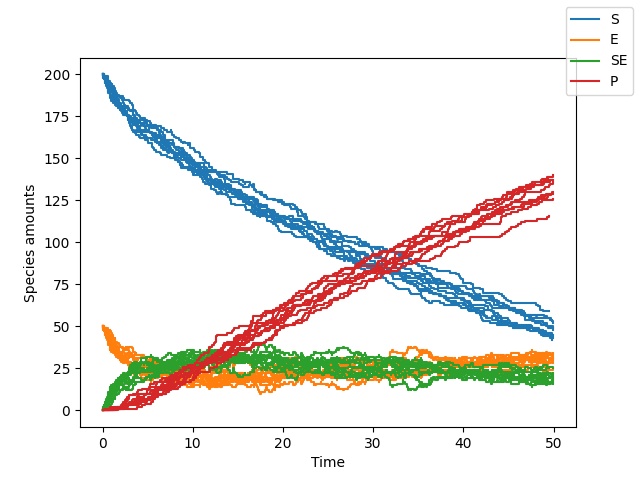
Here we see that the reaction proceeds slower. Less of the product is formed by t=50 compared to the previous case.